Prostate cancer is the second most common cancer affecting men in the world, diagnostic methods and risk assessment remaining a challenge despite decades of research. For years, the specific prostate antigen blood test (PSA) has been a cornerstone in the detection of prostate cancer.
However, although PSA tests reduce deaths related to prostate cancer, this also leads to an over-diagnosis. This requires a change to more individualized and informed approaches to screening, especially for younger men.
The biological role of the PSA goes beyond its use as a diagnostic marker. It plays an essential role in liquefied sperm and tumor progression by interacting with growth factors and proteins in the extracellular matrix. This activity promotes the migration of cancer cells, bone metastases and the formation of blood vessels in tumors.
Despite its clinical usefulness, the PSA’s ability to distinguish between aggressive and indolent cancers remains limited. Recent progress of genetics offer an overview of this limitation and have opportunities for improving diagnostic methods.
Association studies at the genome scale (GWAS) have identified more than 450 nucleotide polymorphisms (SNP) linked to the risk of prostate cancer. These SNPs represent approximately 42.6% of the family risk on prostate cancer in individuals of European ancestry. Among them, a specific SNP in the KLK3 gene – which codes PSA – has drawn significant attention.
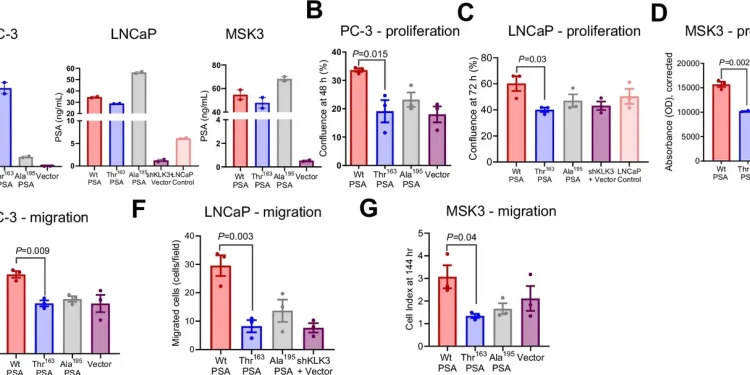
Known as RS17632542, this SNP provokes a substitution of amino acids from isoleucine in Threonine (Ile163thr) in the PSA. This genetic variation is associated with a reduction in the risk of prostate cancer, but its precise role in the disease has remained clear.
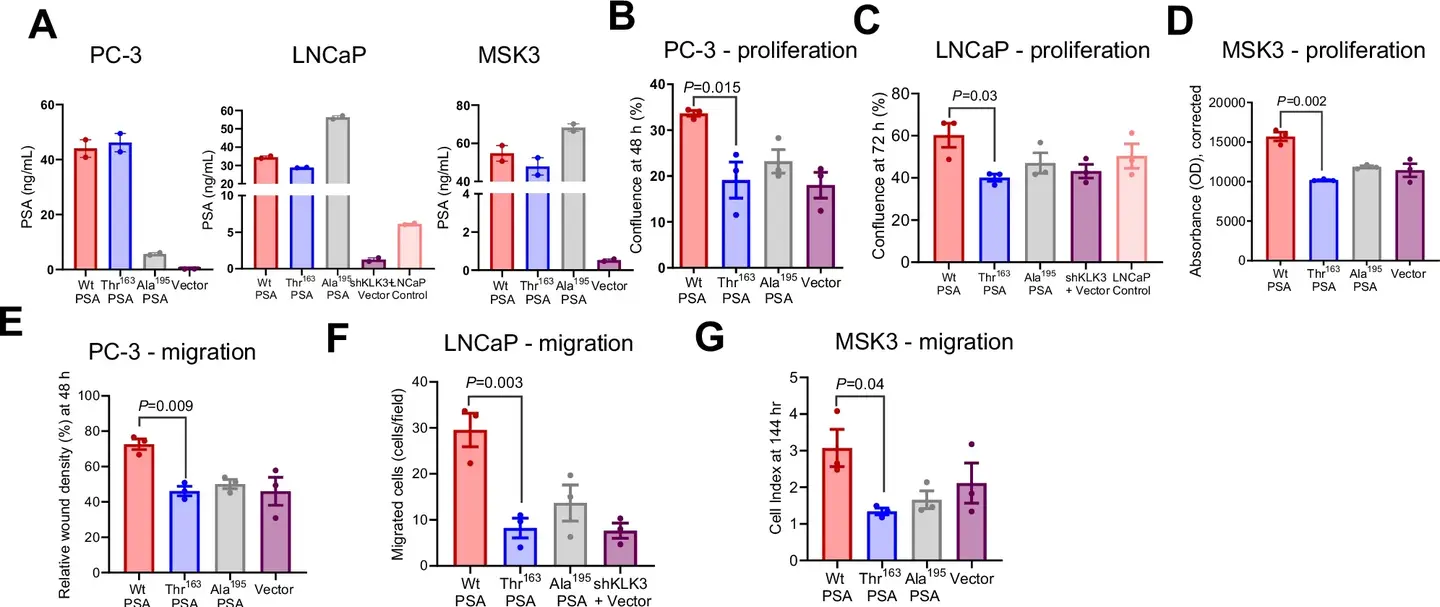
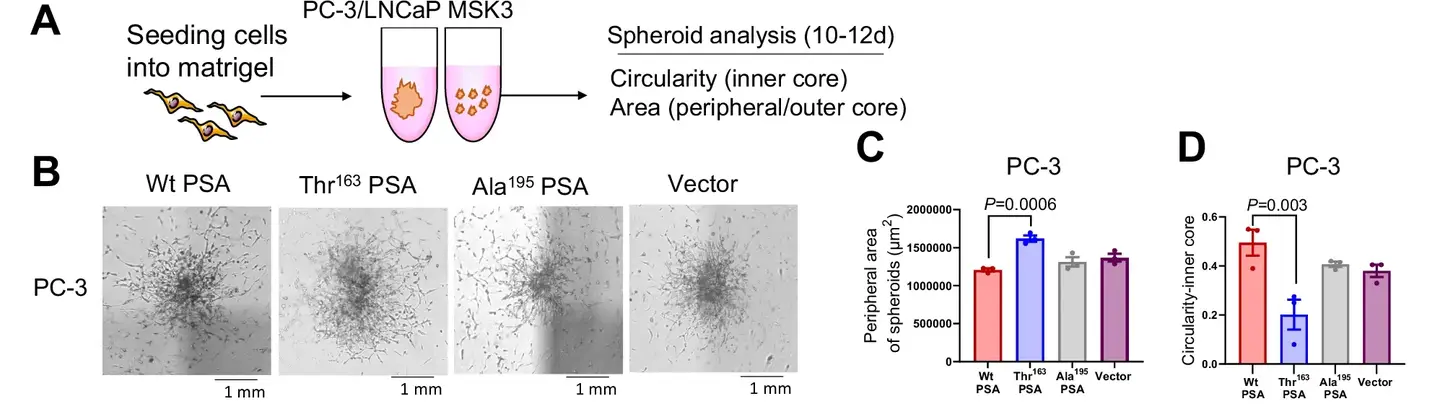
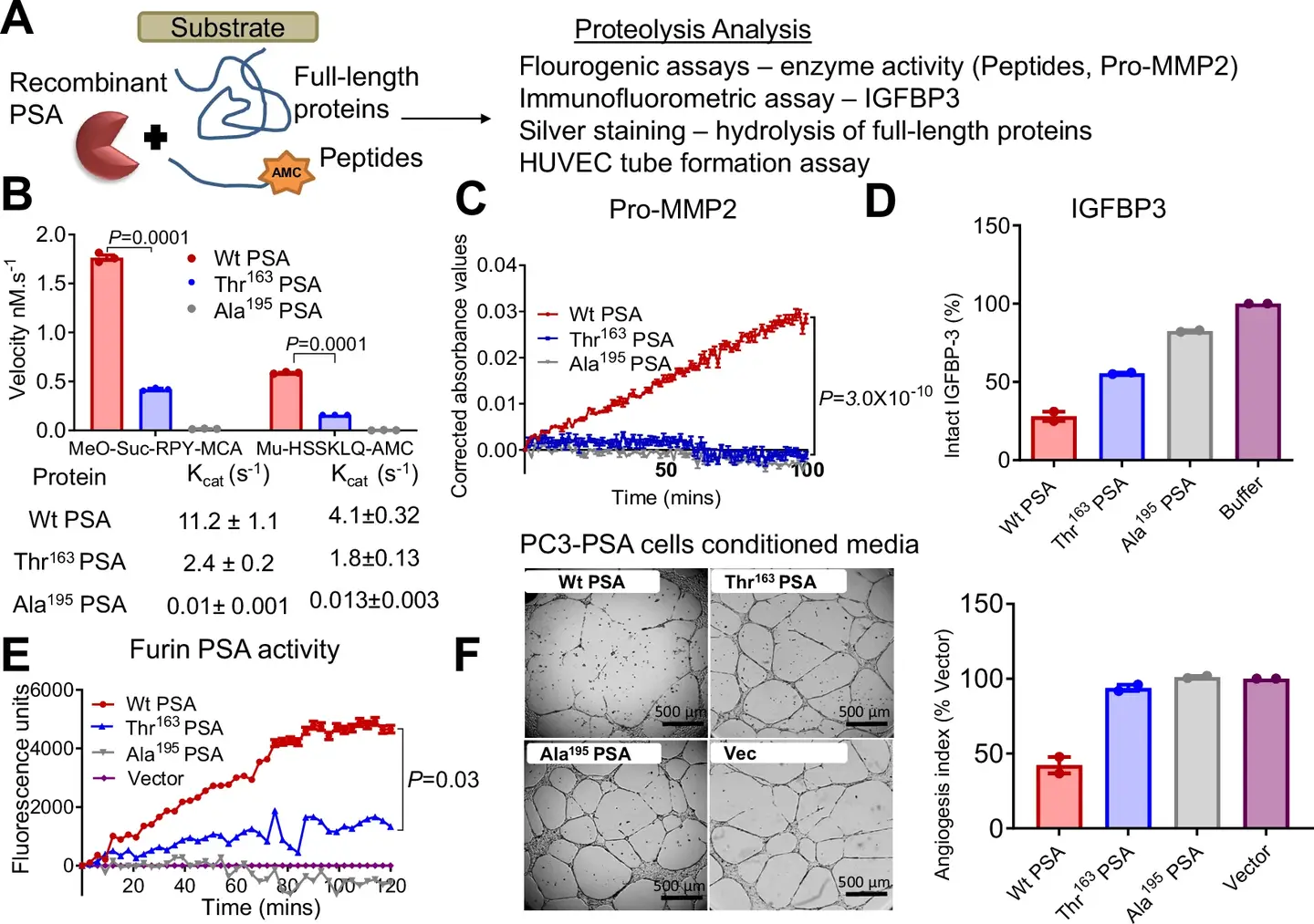
Recent research, published in the journal, Nature Communications, highlight the way in which this SNP influences the biochemical activity of the PSA and its implications for the progression of cancer. Laboratory and animal studies reveal that the Ile163Th variant modifies the proteolytic activity of the PSA, impacting tumor microenvironnement.
This variant is linked to smaller primary tumors but to a higher metastatic potential, in particular to bones, where it triggers the pronounced bone loss. These results highlight the double nature of this genetic variation: although it reduces the overall risk of cancer, it paradoxically increases the probability of an aggressive disease.
The impact of the SNP extends to PSA levels in the blood. The bearers of this variant generally have a lower total PSA and a higher free PSA ratio. This could lead to a delay in diagnosis of prostate cancer, as lower PSA levels may not trigger early biopsy recommendations. Consequently, some aggressive cancers may not be detected to subsequent stages, which causes more worse results.
“Thanks to comprehensive laboratory and mouse tests, we found that this SNP, although associated with a risk of reduced prostate cancer, is also associated with an aggressive type of this cancer,” said Dr. Srinivasan, One of the main researchers in the study. She stressed that the results help explain anomalies in current diagnostic practices and underline the need to improve screening tools.
Despite its limits, the PSA test remains a vital diagnostic tool. However, he cannot make the difference between aggressive and non -aggressive cancers.
This limitation leads to an over-diagnosis, an over-treatment and unnecessary biopsies, which have an impact on the quality of life of patients and increase the costs of health care. At the same time, low PSA levels associated with aggressive cancers can cause delayed diagnosis and higher mortality.
Professor Jyotsna Batra, who led the study, underlined the potential for personalized medicine to meet these challenges. “The results of this study can lead to developing a new simple service point system,” she said.
Such a device could identify patients with genetic variations related to aggressive prostate cancer, even when PSA levels are low. This would allow general practitioners to provide tailor -made diagnostic assessments and more effectively guide clinical decisions.
Researchers’ collaboration efforts have an advanced understanding of how genetic variations influence the results of prostate cancer. The research team, led by Professor Batra and Professor Emeritus emeritus Judith Clements, included experts from bioinformatics, biomedical sciences and clinical practice.
Their results could revolutionize the prognostic prediction by distinguishing more aggressive cancers and by identifying high -risk groups which need early intervention.
This revolutionary research highlights the interaction between genetics and diagnostic markers such as PSA. By identifying the functional implications of the RS17632542 SNP, scientists open the way to more precise and individualized approaches for prostate cancer care. The results of the study highlight the importance of integrating genetic data into the screening and routine processing protocols.
While researchers continue to unravel the complexities of prostate cancer genetics, the objective of personalized medicine is close to reality. With tools informed by genetic variations, clinicians could one day offer more precise and effective care, reducing the over-diagnostic burden while improving results for people with an aggressive disease.
This innovative approach aligns with an era of personalized treatment, where genetic ideas transform disease management such as prostate cancer.